- 微生物及免疫學研究所 教授
- 專長領域: 表觀基因體學、病毒學、癌症免疫、癌症代謝
- 辦公室位置: 生物醫學大樓 306 室 (R306)
- TEL (辦公室): +886-2-2826-7111
- TEL (實驗室): +886-2-2826-7000 ext 65617
- E-mail: pcchang@nycu.edu.tw
- ORCID: 0000-0001-8665-5494

學歷
博士 國立陽明大學 生化暨分子生物研究所
碩士 藥理學研究所 國立陽明大學
經歷
國立陽明大學微生物及免疫學研究所 特聘教授 (2021~2023)
Lerner Research Institute, Cleveland Clinic訪問學者 (2021~迄今)
國立陽明大學微生物及免疫學研究所 教授 (2020~迄今)
國立陽明大學微生物及免疫學研究所 副教授 (2016~2020)
高雄醫學大學傳染病與癌症研究中心 副研究員 (2016~2019)
University of South California (USC), School of Pharmacy 訪問學者 (2015~2016)
國立陽明大學 微生物及免疫學研究所 助理教授 (2011~2016)
獲獎 :
111-114學年度: 績優教研人員
110學年度: 國立陽明交通大學特聘教授
110學年度: 國立陽明交通大學終生免評教師
110學年度: 國立陽明交通大學資深優良教師
110-111學年度: 醫學系網路教學評量 優良教師獎狀
110學年度: 國立陽明交通大學生命科學院第二季重要期刊論文
109學年度: 國立陽明交通大學生命科學院第二季重要期刊論文
108學年度: 國立陽明交通大學醫學系良師益友
105-109學年度: 醫學系網路教學評量 優良教師獎狀
103學年度: 醫學系網路教學評量 優良教師琉璃獎座
102學年度: 國立陽明交通大學生命科學系良師益友導師獎
101-102學年度: 醫學系網路教學評量 優良教師獎狀
研討會主持人/主辦人
The 27th International Workshop on Kaposi’s Sarcoma Herpesvirus and Related Agents (2025/06/29-20255/07/02; California, USA) 研討會主持人
國際邀請演講
University of Pittsburg Medical Center (Aug. 2022)
參訪 2008 年病毒學諾貝爾獎得主 Harald zur Hausen 實驗室 (Jul. 2017)
Seminar serious of USC School of Pharmacy (Feb. 2015)
International Conference on Infectious Diseases and Cancer (Oct. 2014)
The 10th UCSD-UST NYMU-NCTU (Nov. 2016)
The 8th UCSD-UST NYMU-NCTU (Nov. 2014)
論文
Virus-related Publications
- S. Yang, D. Kim, S. Kang, C.J. Lai, I. Cha, P.C. Chang, J.U. Jung. Development of KSHV vaccine platforms and chimeric MHV68-K-K8.1 glycoprotein for evaluating the in vivo immunogenicity and efficacy of KSHV vaccine candidates. mBio 2024; e0291324.
- W. Yeh, Y.Q. Chen, W.S. Yang, Y.C. Hong, S. Kao, T.T. Liu, T.W. Chen, L. Chang, P.C. Chang*. SUMO modification of histone demethylase KDM4A in Kaposi’s sarcoma-associated herpesvirus-induced primary effusion lymphoma. Journal of Virology 2022; 96(16):e0075522.
- S. Yang, W.W. Yeh, M. Campbell, L. Chang, P.C. Chang*. Long non-coding RNA KIKAT/LINC01061 as a novel epigenetic regulator that relocates KDM4A on chromatin and modulates viral reactivation. PLoS Pathogens 2021; 17(6):e1009670.
- Campbell, W.S. Yang, W.W. Yeh, C.H. Kao, P.C. Chang*. Epigenetic Regulation of Kaposi’s sarcoma-associated herpesvirus Latency. Frontiers in Microbiology 2020; 19(11):850.
- W. Liang, M.L. Wang, C.H. Chien, Y.P. Yang, A.A. Yarmishyn, W.Y. Lai, Y.H. Luo, Y.T. Lin, P.C. Chang*, S.H. Chiou*. Highlight of Immune Pathogenic Response and Hematopathologic Effect in SARS-CoV, MERS-CoV, and SARS-Cov-2 Infection. Frontiers in Immunology 2020; 12(11):1022.
- S. Yang, T.Y. Lin, L. Chang, W.W. Yeh, S.C. Huang, T.Y. Chen, Y.T. Hsieh, S.T. Chen, W.C. Li, C.C. Pan, M. Campbell, C.H. Yen, Y.A. Chen, P.C. Chang*. HIV-1 Tat Interacts with a Kaposi’s Sarcoma-Associated Herpesvirus Reactivation-Upregulated Antiangiogenic Long Noncoding RNA, LINC00313, and Antagonizes Its Function. Journal of Virology 2020; 94(3):e01280-19.
- J. Lin, L. Chang, H.W. Chu, H.J. Lin, P.C. Chang, R.Y.L. Wang, B. Unnikrishnan, J.Y. Mao, S.Y. Chen, C.C. Huang. High Amplification of the Antiviral Activity of Curcumin through Transformation into Carbon Quantum Dots. Small 2019; 15(41):e1902641.
- S. Yang, M. Campbell, H.J. Kung, P.C. Chang*. In vitro SUMOylation Assay to Study SUMO E3 Ligase Activity. JoVE-Journal of Visualized Experiments 2018; 131(e56629):1-6.
- S. Yang, M. Campbell, P.C. Chang*. SUMO modification of a heterochromatin histone demethylase JMJD2A enables viral gene transactivation and viral replication. PLoS Pathogens 2017; 13(2):e1006216.
- C. Chang*, M. Campbell, E.S. Robertson. Human Oncogenic Herpesvirus and Post-translational Modifications – Phosphorylation and SUMOylation. Frontiers in Microbiology 2016; 7:962.
- S. Yang, H.W. Hsu, M. Campbell, C.Y. Cheng, P.C. Chang*. K-bZIP mediated SUMO-2/3 specific modification on the KSHV genome negatively regulates lytic gene expression and viral reactivation. PLoS Pathogens 2015; 11(7): e1005051.
- C. Chang* and H.J. Kung. SUMO and KSHV Replication. Cancers 2014; 6(4):1905-1924.
- M. Campbell, K.Y. Kim, P.C. Chang, S. Huerta, B. Shevchenko, D.H. Wang, C. Izumiya, H.J. Kung, Y. A Lytic Viral Long Non-coding RNA Modulates the Function of a Latent Protein. Journal of Virology 2014; 88(3):1843~1848.
- Y. Cheng, C.H. Chu, H.W. Hsu, F.R. Hsu, C.Y. Tang, W.C. Wang, H.J. Kung, P.C. Chang*. An improved ChIP-seq peak detection system for simultaneously identifying post-translational modified transcription factors by combinatorial fusion, using SUMOylation as an example. BMC genomics 2014; 15:S1.
- C. Chang*, C.Y. Cheng, M. Campbell, Y.C. Yang, H.W. Hsu, T.Y. Chang, C.H. Chu, Y.W. Lee, C.L. Hung, S.M. Lai, C.G. Tepper, W.P. Hsieh, H.W. Wang, C.Y. Tang, W.C. Wang, H.J. Kung. The chromatin modification by SUMO-2/3 but not SUMO-1 prevents the epigenetic activation of key immune-related genes during Kaposi’s sarcoma associated herpesvirus reactivation. BMC Genomics 2013; 14(1):824.
- M. Campbell, P.C. Chang, S. Huerta, C. Izumiya, R. Davis, C.G. Tepper, K.Y. Kim, B. Shevchenko, D.H. Wang, J.U. Jung, P.A. Luciw, H.J. Kung, Y. Izumiya. Protein Arginine Methyltransferase 1-directed Methylation of Kaposi Sarcoma-associated Herpesvirus Latency-associated Nuclear Antigen. Journal of Biological Chemistry 2012; 287(8):5806–
- P.C. Chang*, Y. Izumiya, C.Y. Wu, L.D. Fitzgerald, M. Campbell, T. Ellison, K.S. Lam, P.A. Luciw, H.J. Kung. Histone demethylase JMJD2A regulates Kaposi’s sarcoma-associated herpesvirus replication and is targeted by a viral transcriptional factor. Journal of Virology 2011; 85(7):3283-3293.
- P.C. Chang*, L.D. Fitzgerald, D.A. Hsia, Y. Izumiya, C.Y. Wu, W.P. Hsieh, S.F. Lin, M. Campbell, K.S. Lam, P.A. Luciw, C.G. Tepper, H.J. Kaposi’s sarcoma associated herpesvirus (KSHV) encodes a SUMO E3 ligase which is SIM-dependent and SUMO-2/3-specific. Journal of Biological Chemistry 2010; 285(8):5266-5273.
- P.C. Chang*, L. Fitzgerald, A. Van Geelen, Y. Izumiya, T.J. Ellison, D.H. Wang, D.K. Ann, P.A. Luciw, H.J. Kung. KRAB domain-associated protein-1 as a latency regulator for Kaposi’s sarcoma-associated herpesvirus and its modulation by the viral protein kinase. Cancer Research 2009; 69(14):5681-5689.
- Y.L. Lin, P.C. Chang, Y. Wang, and M. Li. Identification of novel viral interleukin-10 isoforms of human cytomegalovirus AD169. Virus Research 2008; 131(2):213-223.
- P. C. Chang* and M. Li. Kaposi’s sarcoma-associated herpesvirus K-cyclin interacts with Cdk9 and stimulates Cdk9-mediated phosphorylation of p53 tumor suppressor. Journal of Virology 2008; 82(1):278-290.
- P. C. Chang*, C.W. Chi, G.Y. Chau, F.Y. Li, Y.H. Tsai, J.C. Wu and Y.H.W. DDX3, a DEAD box RNA helicase, is deregulated in hepatitis virus associated hepatocellular carcinoma and is involved in cell growth control. Oncogene 2006; 25(14):1991-2003.
Cancer-related Publications
- A. Chen, P. C. Chang*, W.W. Yeh, T.Y. Hu, Y.C. Hong, Y.C. Wang, W.J. Huang, T.P. Lin. The lncRNA TPT1-AS1 promotes the survival of neuroendocrine prostate cancer cells by facilitating autophagy. American Journal of Cancer Research 2024 May 15;14(5):2103-2123.
- C. Hong, T.Y. Hu, C.S. Hsu, W.W. Yeh, W.Z. Wong, T.W. Shen, C.H. Chang, K. Hua, C.Y. Tung, Y.C. Peng, C.C., W.J. Huang, P.C. Chang, T.P. Lin. Single-cell analysis of castration-resistant prostate cancers to identify potential biomarkers for diagnosis and prognosis of neuroendocrine prostate cancer. American Journal of Cancer Research 2023 Oct 15;13(10):4560-4578.
- H. Chang, T.Y. Cheng, W.W. Yeh, Y.L. Luo, M. Campbell, T.C. Kuo, T.W. Shen, Y.C. Hong, C.H. Tsai, Y.C. Peng, C.C. Pan, M.H. Yang, J.C. Shih, H.J. Kung, W.J. Huang, P.C. Chang*, T.P. Lin. REST-repressed lncRNA LINC01801 induces neuroendocrine differentiation in prostate cancer via transcriptional activation of autophagy. American Journal of Cancer Research 2023 Sep 15;13(9):3983-4002.
- T. Chang, T.P. Lin, J.T. Tang, M. Campbell, Y.L. Luo, S.Y. Lu, C.P. Yang, T.Y. Cheng, C.H. Chang, T.T. Liu, C.H. Lin, H.J. Kung, C.C. Pan, P.C. Chang*. HOTAIR is a REST-regulated LncRNA that Promotes Neuroendocrine Differentiation in Castration Resistant Prostate Cancer. Cancer Letters 2018; 433:43-52.
- C. Lin, Y.T. Chang, M. Campbell, H.C. Lee, J. C. Shih, P.C. Chang*. MAOA- a novel decision maker of apoptosis and autophagy in hormone refractory neuroendocrine prostate cancer cells. Scientific Reports 2017; 7:46338.
- T. Chang, T.P Lin, M. Campbell, C.C. Pan, S.H. Lee, H.C. Lee, M.H. Yang, H.J. Kung, P.C. Chang*. REST is a crucial regulator for acquiring EMT-like and stemness phenotypes in hormone-refractory prostate cancer. Scientific Reports 2017; 3(7):42795.
- P. Lin, Y.T. Chang, S.Y. Lee, M. Campbell, T.C. Wang, S.H. Shen, H.J. Chung, Y.H. Chang, Allen W Chiu, C.C. Pan, C.H. Lin, C.Y. Chu, H.J. Kung, C.Y. Cheng, P.C. Chang*. REST reduction is essential for hypoxia-induced neuroendocrine differentiation of prostate cancer cells by activating autophagy signaling. Oncotarget 2016; 7:26137~26151.
- C. Chang*, T.Y. Wang, Y.T. Chang, C.Y. Chu, C.L. Lee, H.W. Hsu, T.A. Zhou, Z. Wu, R.H. Kim, S.J. Desai, S. Liu, H.J. Kung. Autophagy Pathway Is Required for IL-6 Induced Neuroendocrine Differentiation and Chemoresistance of Prostate Cancer LNCaP Cells. PLoS ONE 2014; 9:e88556.
- Z. Wu, P.C. Chang, J. Yang, C.Y. Chu, L.Y. Wang, N.T. Chen, A.H. Ma, S.J. Desai, S.H. Lo, C.P. Evans, K.S. Lam, and H.J. Kung. Autophagy Blockade Sensitizes Prostate Cancer Cells towards Src Family Kinase Inhibitors. Genes & Cancer 2010; 1(1):40-49.
Other Publications
- P. Tsao, F.Y. Tseng, C.W. Chao, M.H. Chen, Y.C. Yeh, B.O. Abdulkareem, S.Y. Chen, W.T. Chuang, P.C. Chang, I.C. Chen, P.H. Wang, C.S. Wu, C.Y. Tsai, S.T. Chen. NLRP12 is an innate immune checkpoint for repressing IFN signatures and attenuating lupus nephritis progression. Journal of Clinical Investigation 2023;133(3):e157272.
- T. Hsieh, T.L. Tsai, S.Y. Huang, J.W. Heng, Y.C. Huang, P.Y. Tsai, C.C. Tu, T.L. Chao, Y.M. Tsai, P.C. Chang, C.K. Lee, G.Y. Yu, S.Y. Chang, I.L. Dzhagalov, C.L. Hsu. IFN-stimulated metabolite transporter ENT3 facilitates viral genome release. EMBO Reports 2023; 24(3):e55286.
- C. Tsai, W.H. Chiang, C.H. Wu, Y.C. Li, M. Campbell, P.H. Huang, M.W. Lin, C.H. Lin, S.M. Cheng, P.C. Chang*, C.C. Cheng. miR-548aq-3p is a novel target of Far infrared radiation which predicts coronary artery disease endothelial colony forming cell responsiveness. Scientific Reports 2020; 10(1):6805.
- P. Yang, W.S. Yang, Y.H. Wong, K.H. Wang, Y.C. Teng, M.H. Chang, K.H. Liao, F.S. Nian, C.C. Chao, JW Tsai, W.L. Hwang, M.W. Lin, T.Y. Tzeng, P.N. Wang, M. Campbell, L.K. Chen, T.F. Tsai, P.C. Chang*, H.J. Kung. Muscle atrophy-related myotube-derived exosomal microRNA in neuronal dysfunction: Targeting both coding and long noncoding RNAs. Aging Cell 2020; 19(5):e13107.
- Kant, C.H. Yen, J.H. Hung, C.K. Lu, C.Y. Tung, P.C. Chang, Y.H. Chen, Y.C. Tyan, Y.A. Chen. Induction of GNMT by 1,2,3,4,6-penta-O-galloyl-beta-D-glucopyranoside through proteasome-independent MYC downregulation in hepatocellular carcinoma. Scientific Reports 2019; 9(1):1968.
- H. Liu, J.W. Tsai, J.L. Chen, W.S. Yang, P.C. Chang, P.L. Cheng, D.L. Turner, Y. Yanagawa, T.W. Wang, J.Y. Yu. Ascl1 promotes tangential migration and confines migratory routes by induction of Ephb2 in the telencephalon. Scientific Reports 2017; 7(42895):1.
- F. Chiou, P.C. Chang, C.I. Chou, C.F. Chen. Protein constituent contributes to the hypotensive and vasorelaxant activities of Cordyceps Sinensis. Life Sciences 2000; 66(14):1369-1376.
計畫
- Characterization of TWEAK as a novel circulating aging factor [TWEAK成為循環老化因子之特性研究] PI, 2023/08/01-2026/07/31, National Science and Technology Council (NSTC) (112-2314-B-A49 -023 -MY3; 3 years)
- Study the SUMOylation of histone demethylase KDM4A in the pathogenesis and treatment of KSHV-associated primary effusion lymphoma (PEL) [探討 SUMO 修飾組織蛋白去甲基酶KDM4A 對卡波西肉瘤相關性皰疹病毒感染所引發的原發性積液淋巴瘤之致病機制與治療潛力] PI, 2022/08/01-2025/07/31, National Science and Technology Council (NSTC) (111-2320-B-A49 -030 -MY3; 3 years)
- Investigation of the mechanism and medical implications of a novel histone lysine demethylase KDM4A interacting long non-coding RNAs (LncRNAs) LINC01061 in viral reactivation and tumorigenesis [探討組蛋白去甲基酶 KDM4A 結合長鏈非編碼RNA (lncRNAs) LINC01061 參與病毒活化與細胞癌化的機制及其醫療角色(111-113)] PI, 2022/01/01-2024/12/31, National Health Research Institutes (NHRI-EX111-11125BI; 3 years)
- Epigenetic regulation of IL-10 production in virus induced B-lymphocyte in modulation host inflammatory response and cell pathogenesis[探討病毒誘發B淋巴細胞生成IL-10之表關基因體調控及調節宿主細胞之發炎反應(1/2~2/2)] PI, 202107-201312, Ministry of Science and Technology (MOST) (109-2926-I-010-503 -, 111-2926-I-010-502-G; 2 years) — 與CCF合作計畫
- Using KSHV reactivation as a model to identify oncogenic long non-coding RNAs (LncRNAs) and elucidate their role in epigenetic regulation of human cancer [利用致癌性卡波氏肉瘤相關性皰疹病毒當研究模式尋找癌化相關之長鏈非編碼RNA並探討其在表觀遺傳性腫瘤發生過程中所扮演的角色] PI, 2019/08/01-2022/07/31, Ministry of Science and Technology (MOST) (108-2320-B-010-029-MY3; 3 years)
- Using KSHV as a model to study the role of SUMOylation in regulating the chromatin remodeling mediated transcription regulation and oncogenic characteristic of JMJD2A [利用卡波西肉瘤相關性皰疹病毒當作實驗模式:探討SUMO修飾組織蛋白去甲基酶JMJD2A對其附基因體調控基因表現與細胞癌化能力之影響] PI, 20160801-20190731, Ministry of Science and Technology (MOST) (105-2320-B-010-007-MY3; 3 years)
- The role of REST in epigenetic regulation of neuroendocrine cancer stem cell-mediated chemoresistence [REST所調控之附基因體轉化對神經內分泌分化癌症幹細胞導致之攝護腺癌化療機制之研究] PI, 20150801-20160731, Ministry of Science and Technology (MOST) (104-2320-B-010-038-; 1 year)
- MAOA mediates REST action on neuroendocrine differentiation and autophagy activation of prostate cancer relapse[MAOA 在復發型攝護腺癌中REST造成之癌細胞神經內分泌樣分化與自噬機制活化中所扮演的角色(1/2~2/2)] PI, 20140901 ~20170331, Ministry of Science and Technology (MOST) (103-2911-I-010-515 -, 104-2911-I-010-505 ; 2 years) — 與USC合作計畫
- SUMO and KSHV: Epigenetic regulation of Oncogenic Herpesvirus[SUMO與卡波西肉瘤相關皰疹病毒:附基金體調控致癌性病毒皰疹(102-105)] PI, 20130101-20161231, National Health Research Institutes (NHRI-EX105-10215BC; 4 years)
- The role of SUMO-2/3 in epigenetic regulation of herpesvirus: Implication in developing novel SUMO-2/3 target therapy [附基因體模式探式SUMO-2/3 調控疱疹病毒的生長與致病機制:發展SUMO-2/3專一性標靶藥物(1/3~3/3)] PI, 20120801~20150731, National Science Council (NSC) (101-2321-B-010-017-; 102-2321-B-010-015-; 103-2321-B-010-006-; 3 years)
- Autophagy, neuroendocrine differentiation and prostate cancer therapeutic resistance [細胞自噬機制在神經內分泌分化導致之抗攝護腺癌治療作用之探討] PI, 20120801~20150731, National Science Council (NSC) (101-2320-B-010-047-MY3; 3 years).
研究方向
(I) Virus-Host interaction: Epigenetic regulation of herpesvirus oncogenesis
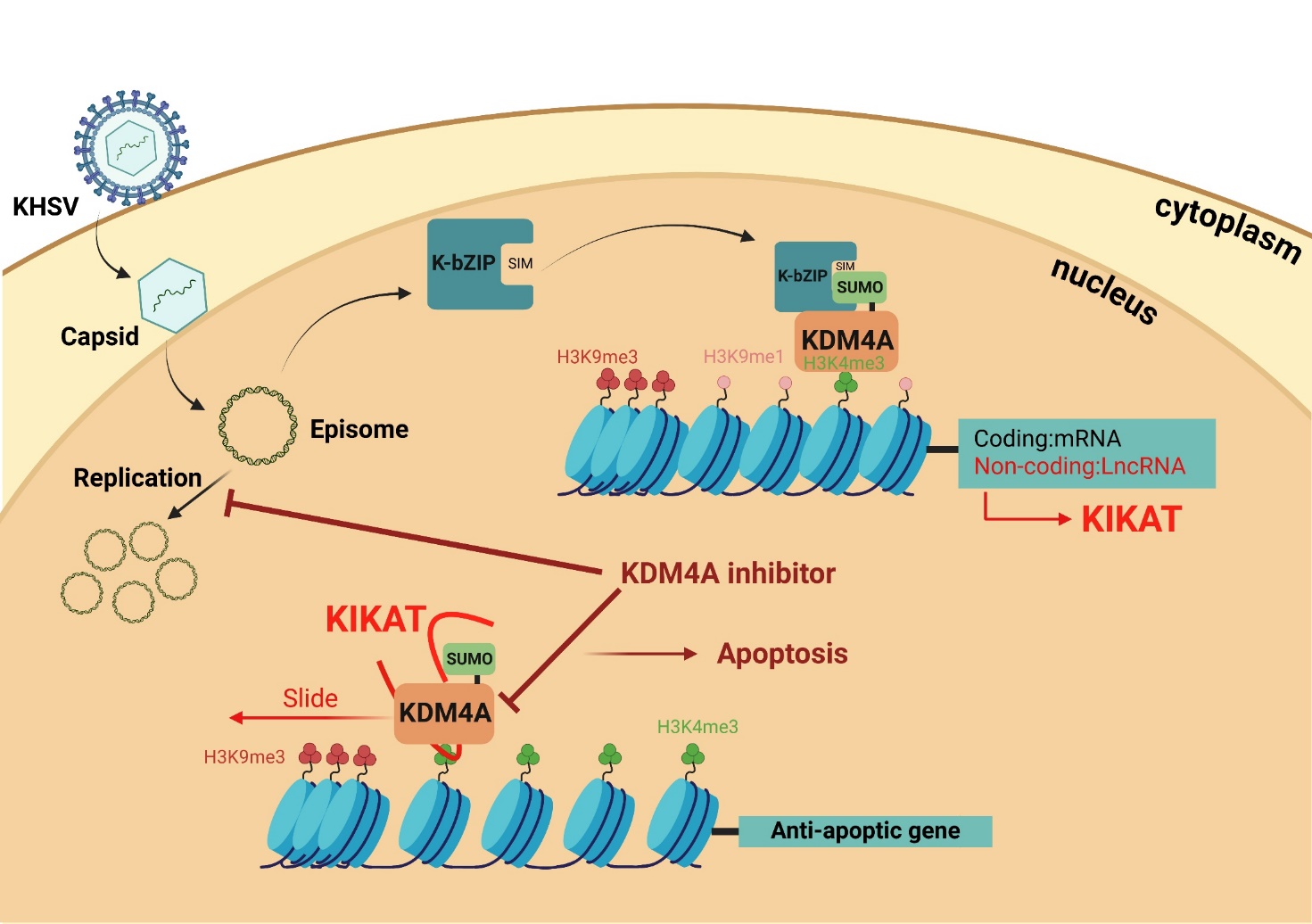
表觀遺傳調控主要分為三大類:
(1) DNA甲基化; (2)組蛋白甲基化; (3)長鏈非編碼RNA。
我們以致癌皰疹病毒KSHV作為研究模型,探討第一個被發現能移除組蛋白三甲基的去甲基酶KDM4A/JMJD2A在表觀遺傳調控與腫瘤形成中的角色。研究發現,KDM4A能讓染色質維持低的基因抑制標誌3K9me3,讓基因在受到刺激時可以快速啟動。進一步發現,KSHV的病毒蛋白K-bZIP,是SUMO E3連接酶,他會對 KDM4A進行SUMO修飾。這個修飾對 KDM4A 結合染色質並發揮去甲基化至關重要。更有趣的是,我們發現KSHV 也會誘導KIKAT新型長鏈非編碼RNA (lncRNA) 表現,KIKAT能引導KDM4A往啟動子區域上游移動,離開轉錄起始位點 (TSS),並形成一個「隔離器」,阻擋H3K9me3入侵,確保基因能快速啟動。此外,我們利用single-cell RNA-seq與bulk RNA-seq證實,KDM4A對KSHV在裂解期的病毒基因表達與病毒產生至關重要,並且能促進感染細胞的存活、移動和血管新生,為病毒提供充足養分。
簡單來說,病毒不只能讓自己在需要時快速再活化並大量複製,還會利用高超的表觀遺傳手法改造細胞染色質環境,「躲避」免疫系統以及導致癌症形成。因此,如果能阻斷KDM4A的功能,就有機會清除感染細胞、抑制癌細胞擴散,且減少病毒量。目前,我們正在深入探索KDM4A在不同病毒與癌症中的角色,並發展以阻斷KDM4A及其SUMO修飾為核心的全新抗病毒致癌治療策略。
(II) Tumor-Immune Interplay in HBV-Associated HCC
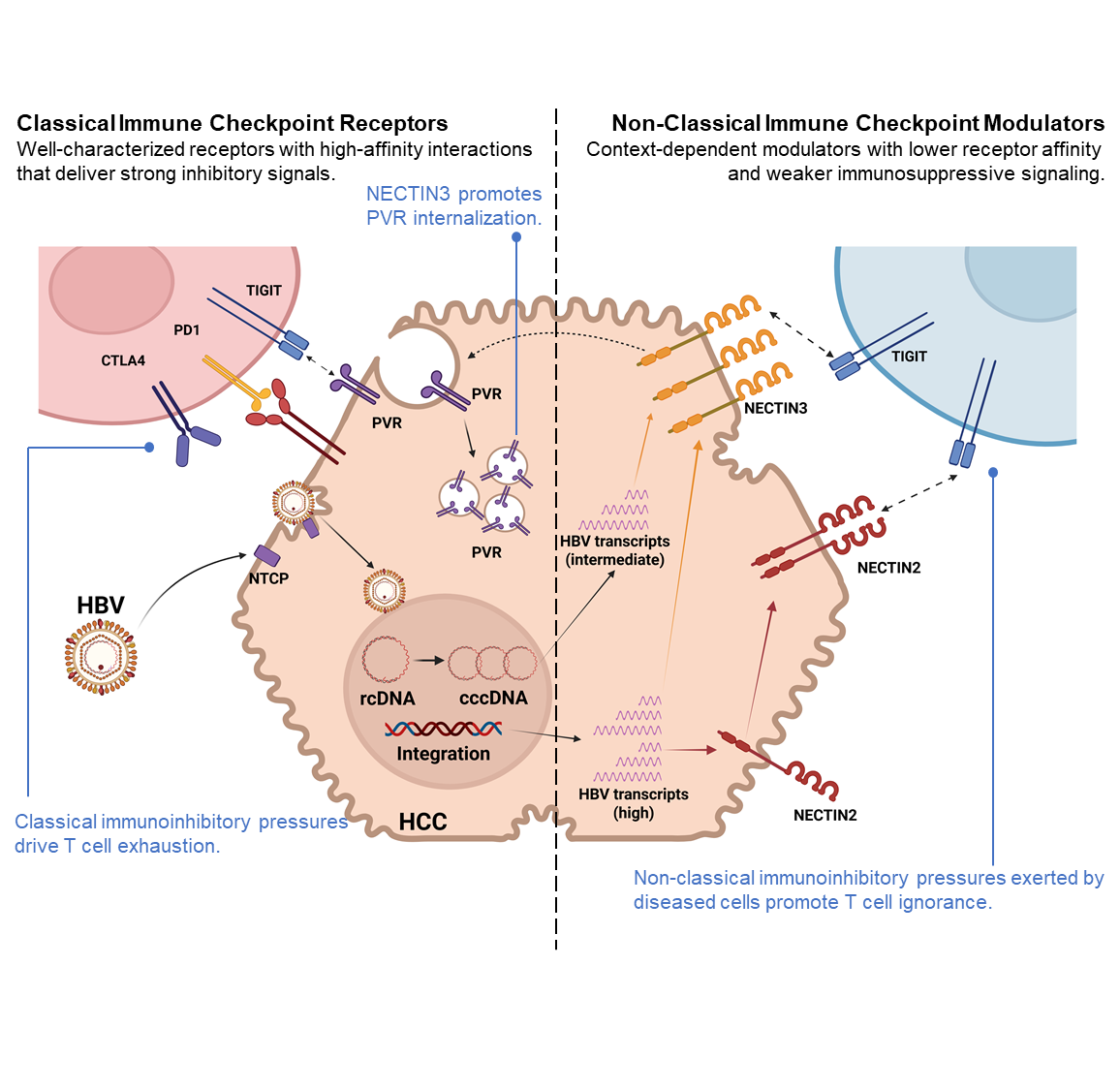
免疫檢查點抑制劑 (ICI) 透過恢復免疫力,重塑癌症治療,但其效果常受T細胞衰竭限制。最新研究發現,T細胞可能進入一種幹細胞狀態,維持T細胞功能,延續免疫效能。病毒相關肝細胞癌 (HCC) 對ICI的反應優於非病毒性HCC,因此我們正探討HBV相關HCC中是否存在具有幹細胞特性之T細胞,並評估其作為免疫療法新標靶的潛力。
(III) Epigenetic regulation of lineage plasticity in prostate cancer progression
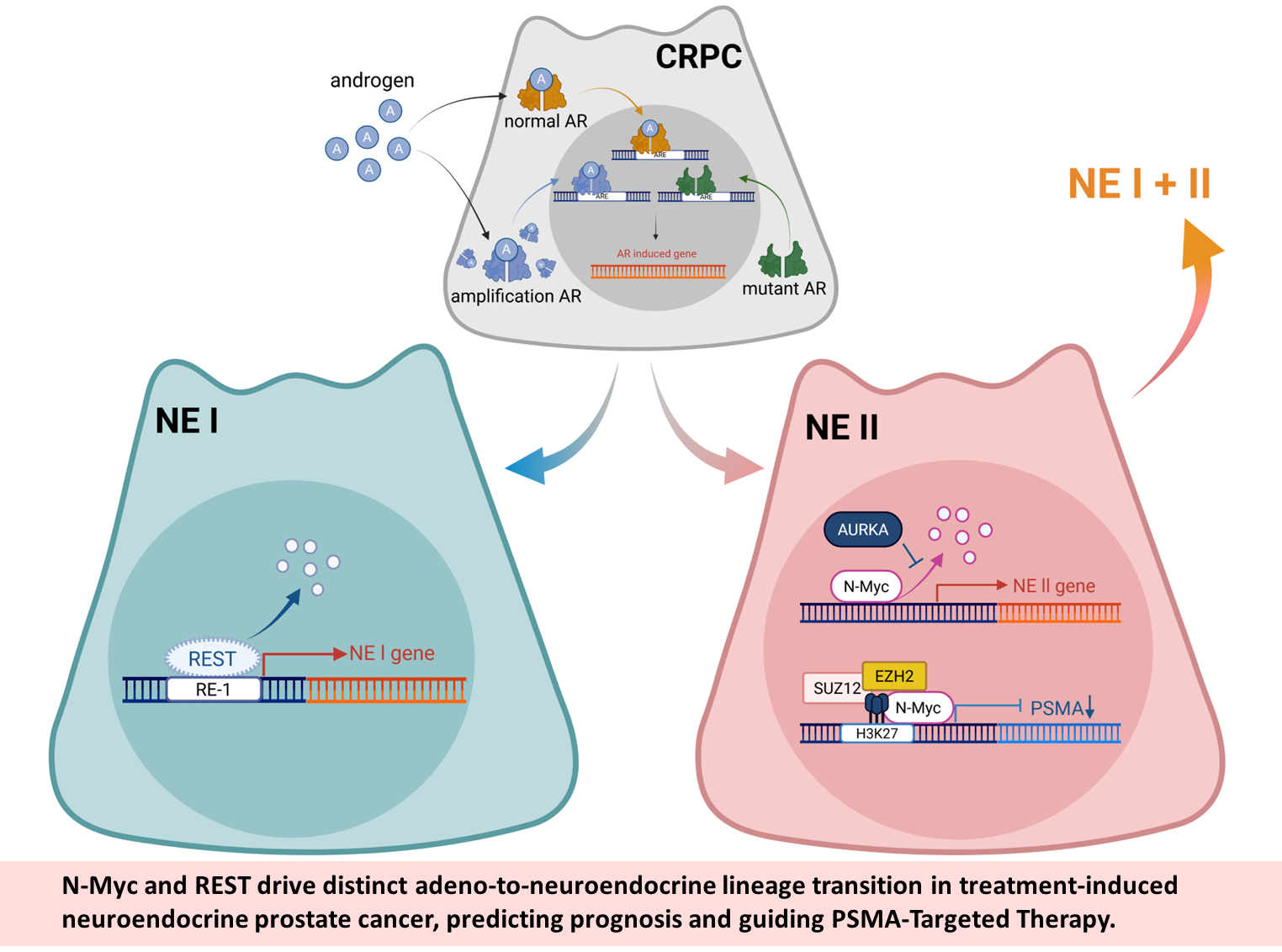
表觀遺傳機制會調控細胞可塑性,能使相同基因型在環境刺激下產生細胞多樣性,並成為治療抗性的關鍵因素。我們的研究發現雄激素剝奪、IL-6 及缺氧可誘導攝護腺癌走向神經內分泌分化。透過RNA-seq與ChIP-seq分析,我們鑑定出多個與NEPC 發展相關的編碼與非編碼基因,經由活化自噬途徑促進腫瘤細胞分化與存活。透過single-cell RNA-seq分析,揭示NEPC具有不同亞群,且發現REST與N-MYC是調控NEPC進入不同亞群的關鍵表觀控因子。這些發現不僅加深我們對致死性攝護腺癌的理解,也為個人化精準醫療提供了新契機。
(IV) Tumor microenvironment in head and neck cancer progression
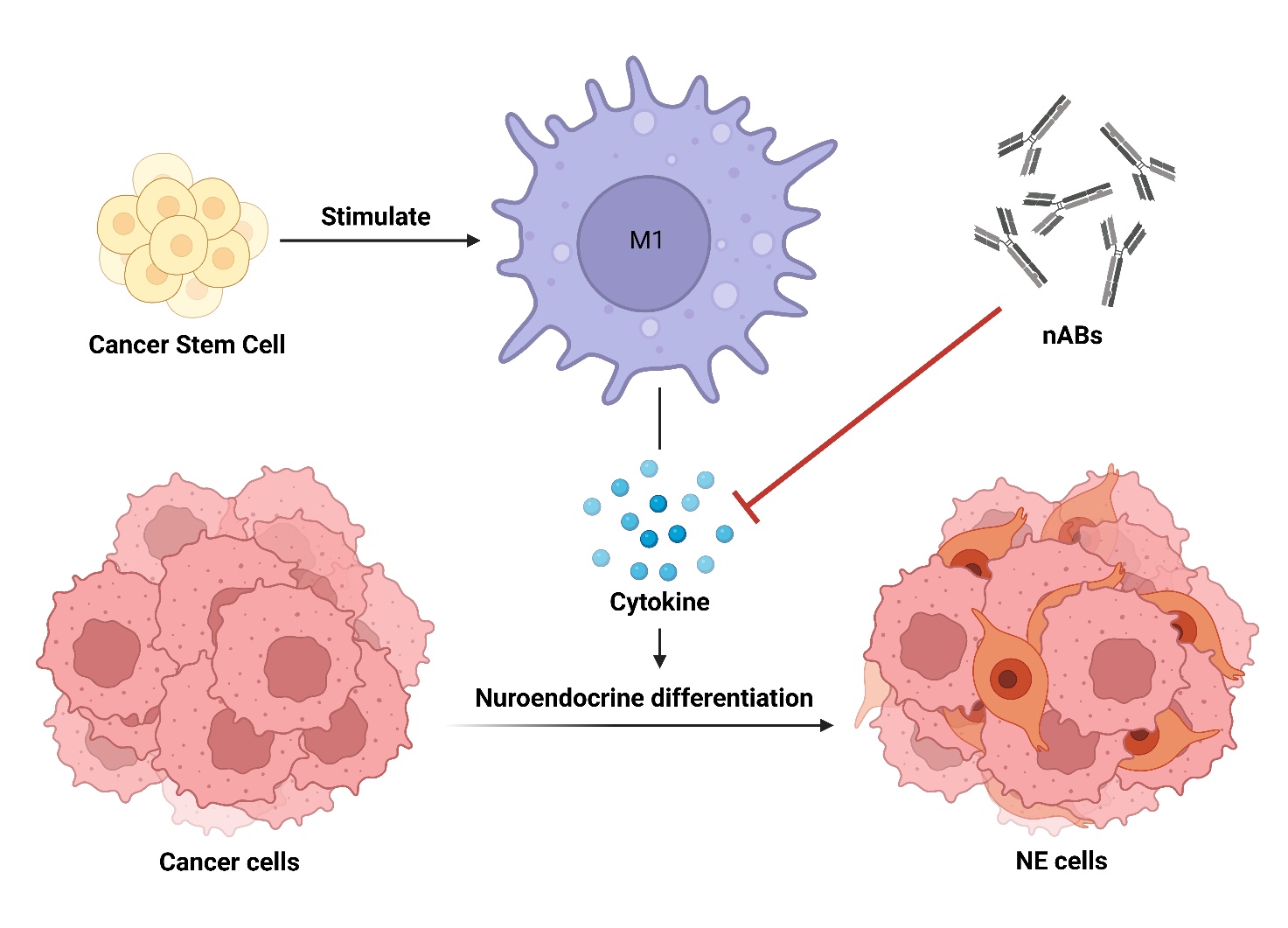
發炎與抗發炎型巨噬細胞所塑造的微環境,不僅參與宿主防禦,也促進癌症的發展。我們發現,癌症幹細胞 (CSCs)會誘導 M1 巨噬細胞產生與神經內分泌分化相關的細胞激素。結合轉錄體與質譜分析,發現醣解作用對於相關細胞激素的產生有重要貢獻。因此,CSC誘導的 M1巨噬細胞會使癌細胞走向治療抗性神經內分泌分化癌。這些發現提供了使用免疫療法來阻止癌症更惡性的治療機會。
(V) Muscle-to-brain axis in aging
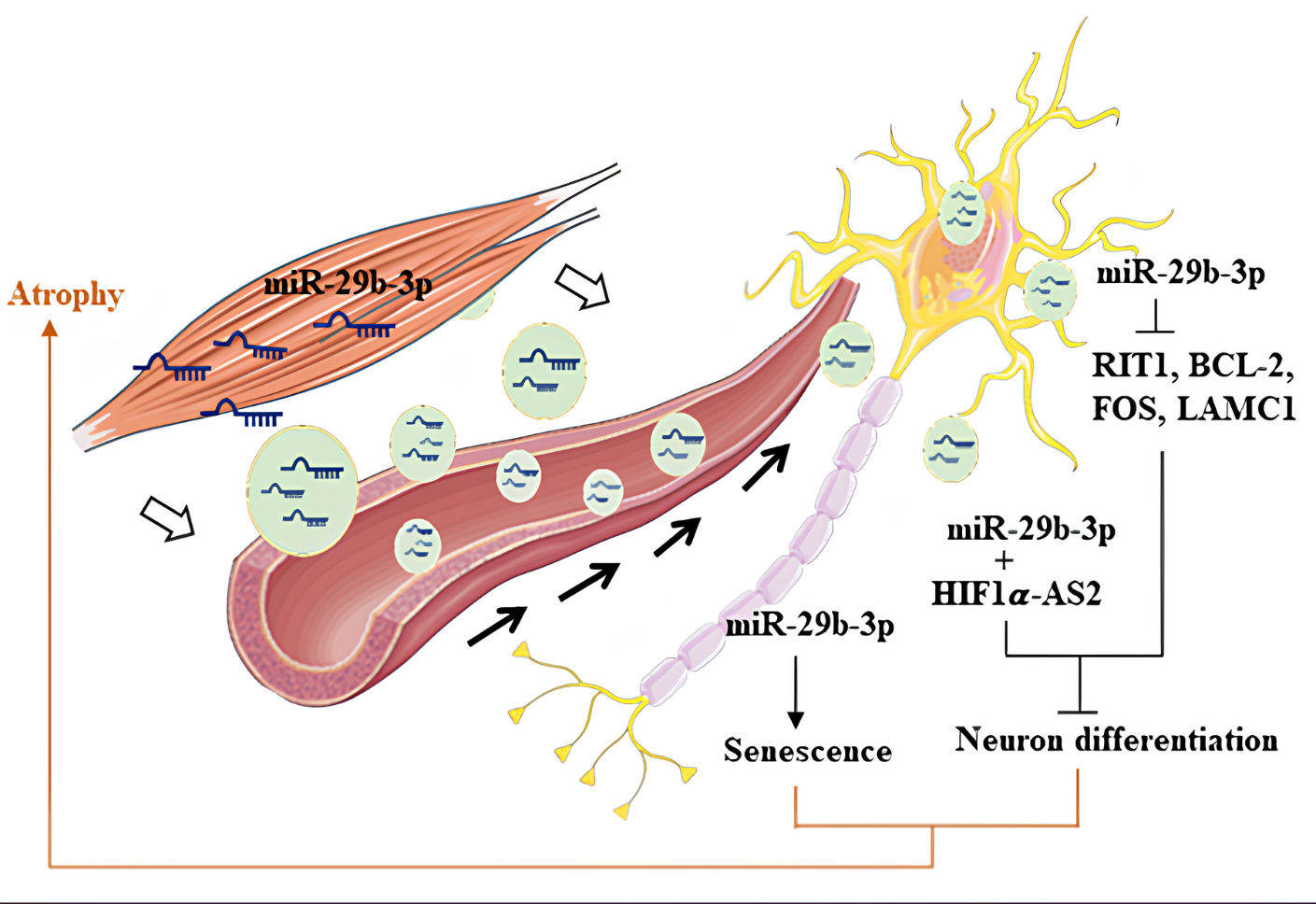
肌肉質量和力量的喪失,稱為肌少症,是老年人虛弱的主要原因,也是生活品質下降和醫療支出增加的主要因素。然而,對於肌肉質量喪失與神經退化之間的關聯尚不清楚。細胞外囊泡 (EVs),例如外泌體,是由細胞釋放的小顆粒,介導細胞間的通訊,將物質轉移到受體細胞。我們的研究發現,來自萎縮肌肉細胞的外泌體,可以經由血液將微小RNA轉移到神經細胞,調控神經細胞中的編碼和非編碼RNA,導致神經細胞功能障礙。
實驗室成員
現在成員:
博士生: 沈曉津
碩士生: 黃儀綸、顏之宸、荊羽芊、吳柏毅
大學部: 王紹丞、楊睿奇、王琮賢
助理: 謝蕙如、黃璵帆
歷屆碩士畢業生 (入學):
2011/09 張怡婷 (論文發表: PLoS One. 2014 Feb 14; 9(2):e88556)
2011/09 楊奕程 (論文發表: BMC Genomics. 2013 Nov 23; 14:824)
2013/09 楊宛珊 (論文發表: PLoS Pathogens. 2015 Jul 21; 11(7):e1005051)
2013/09 李松遠 (論文發表: Oncotarget. 2016 May 3; 7(18):26137)
2015/09 羅勻里
2015/09 黃詩晴
2016/09 楊佳蓓 (論文發表: Aging cell. 2020 Mar 31:e13107 )
2016/09 李悅誠
2017/09 林庭宇 (論文發表: J Virol. 2020 Jan 17;94(3): e01280-19)
2017/09 江玟慧 (論文發表: Sci Rep. 2020 Apr 22;10(1):6805)
2017/09 鄭庭伃 (論文發表: Am J Cancer Res. 2023 Sep 15;13(9):3983-4002)
2017/09 葉 葳 (論文發表: J Virol 2022; 96(16):e0075522.)
2017/09 施文仁
2018/09 高晨媗
2019/09 沈彩文
2020/09 胡則勻 (論文發表: Am J Cancer Res. 2023 Nov;13(10):4560-45782)
2020/09 陳柏安 (論文發表: Am J Cancer Res. 2024 May 15;14(5):2103-2123)
2021/09 鄭裕弘
2022/09 游凱婷
2022/09 簡沛芹
2023/09 洪詠智
歷屆博士畢業生:
張怡婷 (Sep 2014 ~ Jun 2018)
- C. Chang, T.Y. Wang, Y.T. Chang, C.Y. Chu, C.L. Lee, H.W. Hsu, T.A. Zhou, Z. Wu, R.H. Kim, S.J. Desai, S. Liu, H.J. Kung. Autophagy Pathway Is Required for IL-6 Induced Neuroendocrine Differentiation and Chemoresistance of Prostate Cancer LNCaP Cells. PLoS ONE 2014; 9:e88556 .
- P. Lin, Y.T. Chang, S.Y. Lee, M. Campbell, T.C. Wang, S.H. Shen, H.J. Chung, Y.H. Chang, Allen W Chiu, C.C. Pan, C.H. Lin, C.Y. Chu, H.J. Kung, C.Y. Cheng, P.C. Chang. REST reduction is essential for hypoxia-induced neuroendocrine differentiation of prostate cancer cells by activating autophagy signaling. Oncotarget 2016; 7:26137~26151.
- T. Chang, T.P Lin, M. Campbell, C.C. Pan, S.H. Lee, H.C. Lee, M.H. Yang, H.J. Kung, P.C. Chang. REST is a crucial regulator for acquiring EMT-like and stemness phenotypes in hormone-refractory prostate cancer. Scientific Reports 2017; 3(7):42795.
- C. Lin, Y.T. Chang, M. Campbell, H.C. Lee, J. C. Shih, P.C. Chang. MAOA- a novel decision maker of apoptosis and autophagy in hormone refractory neuroendocrine prostate cancer cells. Scientific Reports 2017; 7:46338. [第一作者]
- T. Chang, T.P. Lin, J.T. Tang, M. Campbell, Y.L. Luo, S.Y. Lu, C.P. Yang, T.Y. Cheng, C.H. Chang, T.T. Liu, C.H. Lin, H.J. Kung, C.C. Pan, P.C. Chang. HOTAIR is a REST-regulated LncRNA that Promotes Neuroendocrine Differentiation in Castration Resistant Prostate Cancer. Cancer Letters 2018; 433:43-52.
楊宛珊 (Sep 2015 ~ Jun 2020) (現職: Lerner Research Institute, Cleveland Clinic 博士後研究員)
- S. Yang, W.W. Yeh, M. Campbell, L. Chang, P.C. Chang. Long non-coding RNA KIKAT/LINC01061 as a novel epigenetic regulator that relocates KDM4A on chromatin and modulates viral reactivation. PLoS Pathogens 2021; 17(6):e1009670.
- Campbell, W.S. Yang, W.W. Yeh, C.H. Kao, P.C. Chang. Epigenetic Regulation of Kaposi’s sarcoma-associated herpesvirus Latency. Frontiers in Microbiology 2020; 19(11):850.
- P. Yang, W.S. Yang, Y.H. Wong, K.H. Wang, Y.C. Teng, M.H. Chang, K.H. Liao, F.S. Nian, C.C. Chao, JW Tsai, W.L. Hwang, M.W. Lin, T.Y. Tzeng, P.N. Wang, M. Campbell, L.K. Chen, T.F. Tsai, P.C. Chang, H.J. Kung. Muscle atrophy-related myotube-derived exosomal microRNA in neuronal dysfunction: Targeting both coding and long noncoding RNAs. Aging Cell 2020; 19(5):e13107.
- S. Yang, T.Y. Lin, L. Chang, W.W. Yeh, S.C. Huang, T.Y. Chen, Y.T. Hsieh, S.T. Chen, W.C. Li, C.C. Pan, M. Campbell, C.H. Yen, Y.A. Chen, P.C. Chang*. HIV-1 Tat Interacts with a Kaposi’s Sarcoma-Associated Herpesvirus Reactivation-Upregulated Antiangiogenic Long Noncoding RNA, LINC00313, and Antagonizes Its Function. Journal of Virology 2020; 94(3):e01280-19.
- S. Yang, M. Campbell, H.J. Kung, P.C. Chang. In vitro SUMOylation Assay to Study SUMO E3 Ligase Activity. JoVE-Journal of Visualized Experiments 2018; 131(e56629):1-6.
- S. Yang, M. Campbell, P.C. Chang*. SUMO modification of a heterochromatin histone demethylase JMJD2A enables viral gene transactivation and viral replication. PLoS Pathogens 2017; 13(2):e1006216.
- S. Yang, H.W. Hsu, M. Campbell, C.Y. Cheng, P.C. Chang. K-bZIP mediated SUMO-2/3 specific modification on the KSHV genome negatively regulates lytic gene expression and viral reactivation. PLoS Pathogens 2015; 11(7):e1005051
張龍 (Sep 2016 ~ Feb 2020)
- Lin, L. Chang, H.W. Chu, H.J. Lin, P.C. Chang, R.Y.L. Wang, B. Unnikrishnan, J.Y. Mao, S.Y. Chen, C.C. Huang. High Amplification of the Antiviral Activity of Curcumin through Transformation into Carbon Quantum Dots. Small 2019; 15(41):e1902641. [第一作者]
張景欣 (Sep 2016 ~ July 2024)
- H. Chang,, T.Y. Cheng, W.W. Yeh, Y.L. Luo, M. Campbell, T.C. Kuo, T.W. Shen, Y.C. Hong, C.H. Tsai, Y.C. Peng, C.C. Pan, M.H. Yang, J.C. Shih, H.J. Kung, W.J. Huang, P.C. Chang*, T.P. Lin*. REST-repressed lncRNA LINC01801 induces neuroendocrine differentiation in prostate cancer via transcriptional activation of autophagy. American Journal of Cancer Research 2023; 15;13(9):3983-4002.
- H. Chang, P.H. Yu, P.F. Hsieh, J.H. Hong, C.H. Chiang, H.M. Cheng, H.C. Wu, C.Y. Huang, T.P. Lin. Prostate health index density aids the diagnosis of prostate cancer detected using magnetic resonance imaging targeted prostate biopsy in Taiwanese multicenter study. Journal of the Chinese Medical Association 2024; 1;87(7):678-685.



